In the soils of Eurasia, among bones brittle with age, lies not just the story of ancient humans but also the footprints of the pathogens that plagued them. A recent study led by researchers at the University of Copenhagen and the University of Cambridge has offered one of the most extensive genetic reconstructions of the ancient microbial world to date. Published in Nature1, the study analyzes ancient DNA from more than 1,300 prehistoric individuals and identifies 214 distinct human pathogens, painting a portrait of disease evolution across 37,000 years of human history.
These findings underscore how major shifts in lifestyle—especially the transition to agriculture and animal domestication—reshaped not only the genetic landscape of humans, but also that of their microbial adversaries.
When Livestock Invited Infections
The genomic record suggests that the rise of zoonotic diseases—those transmitted from animals to humans—was tightly coupled to the Neolithic turn. Around 6,500 years ago, as humans clustered into villages and brought livestock into their homes and fields, the earliest clear evidence of animal-borne diseases began to emerge. By 5,000 years ago, these diseases appear to have intensified and diversified.
“We’ve long suspected that the transition to farming and animal husbandry opened the door to a new era of disease—now DNA shows us that it happened at least 6,500 years ago,” said Eske Willerslev, senior author of the study.
The findings come from the painstaking extraction and sequencing of ancient DNA from skeletal remains, revealing not just the people of the past, but their invisible co-travelers: bacteria, viruses, and parasites that once lived inside them.
A Genetic Archive of Plague and Pestilence
Among the discoveries is the oldest known genetic trace of Yersinia pestis, the bacterium responsible for the plague. Detected in a 5,500-year-old individual, this early presence predates the infamous medieval Black Death by several millennia. While the later pandemics are etched into historical record, this ancient strain offers a deeper context: the roots of pandemic pathogens stretch back into the time of early cities, long before the age of written texts.
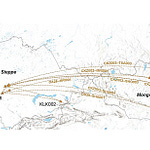
Other pathogens mapped in the study include early forms of hepatitis B, herpesviruses, and parasites that still trouble humans today. The data show not only which diseases were present but how they moved. The migration of Steppe pastoralists across Eurasia, for instance, is correlated with a surge in pathogen diversity, suggesting that people on the move brought their microbes with them.
“These infections didn’t just cause illness—they may have contributed to population collapse, migration, and genetic adaptation,” said Willerslev.
Evolutionary Legacies of Illness
Pathogens do more than make people sick. They shape genomes, carve out survival strategies, and leave marks that persist across generations. The immune systems of contemporary humans still bear traces of these ancient microbial encounters.
Martin Sikora, the study’s first author, emphasized the long arc of these evolutionary interactions. Understanding the spatiotemporal distribution of pathogens may help explain patterns of resistance and susceptibility that persist today.
“If we understand what happened in the past, it can help us prepare for the future,” said Sikora. “Mutations that were successful in the past are likely to reappear.”
In a world where emerging diseases are again top of mind, this research offers a sobering reminder: pandemics are not new. What’s different now is how much more we know—and what we might choose to do with that knowledge.
Implications for Today and Tomorrow
There is hope that ancient pathogens can inform modern medicine. By identifying how past microbial threats evolved, scientists may improve the design of vaccines and better predict how pathogens might mutate. A genetic mutation that once helped a pathogen evade the immune system thousands of years ago might still be effective today, hiding in the viral toolbox, ready to resurface.
In this way, the ancient genome acts not only as a window into past health, but as a diagnostic lens through which the threats of the future might be glimpsed.
Related Research
Andrades Valtueña, A., et al. (2017). The Stone Age plague: 1000 years of persistence in Eurasia. Cell, 173(6), 1357–1364.e14. https://doi.org/10.1016/j.cell.2018.03.042
Ancient Yersinia pestis genomes from the Neolithic and Bronze Age show early forms of plague before major pandemics.
Rasmussen, S., et al. (2015). Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell, 163(3), 571–582. https://doi.org/10.1016/j.cell.2015.10.009
Demonstrates how early plague strains circulated widely before becoming pandemic-causing.
Spyrou, M. A., et al. (2019). A European Yersinia pestis lineage from the second plague pandemic. Nature Communications, 10, 4470. https://doi.org/10.1038/s41467-019-12154-0
Illuminates later-stage plague evolution through medieval genomes.
Key, F. M., et al. (2020). Emergence of human-adapted Salmonella enterica is linked to the Neolithization process. Nature Ecology & Evolution, 4, 324–333. https://doi.org/10.1038/s41559-019-1060-1
Connects Neolithic farming transitions to emergence of human-adapted pathogens.
Houldcroft, C. J., & Underdown, S. J. (2016). Neanderthal genomics and the evolution of modern human disease susceptibility. Journal of Molecular Evolution, 82(4-5), 192–203. https://doi.org/10.1007/s00239-016-9732-0
Explores how interbreeding with Neanderthals influenced immune responses and disease.
Sikora, M., Canteri, E., Fernandez-Guerra, A., Oskolkov, N., Ågren, R., Hansson, L., Irving-Pease, E. K., Mühlemann, B., Holtsmark Nielsen, S., Scorrano, G., Allentoft, M. E., Valeur Seersholm, F., Schroeder, H., Gaunitz, C., Stenderup, J., Vinner, L., Jones, T. C., Nystedt, B., Sjögren, K.-G., … Willerslev, E. (2025). The spatiotemporal distribution of human pathogens in ancient Eurasia. Nature, 1–9. https://doi.org/10.1038/s41586-025-09192-8