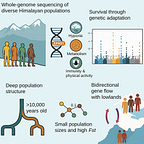
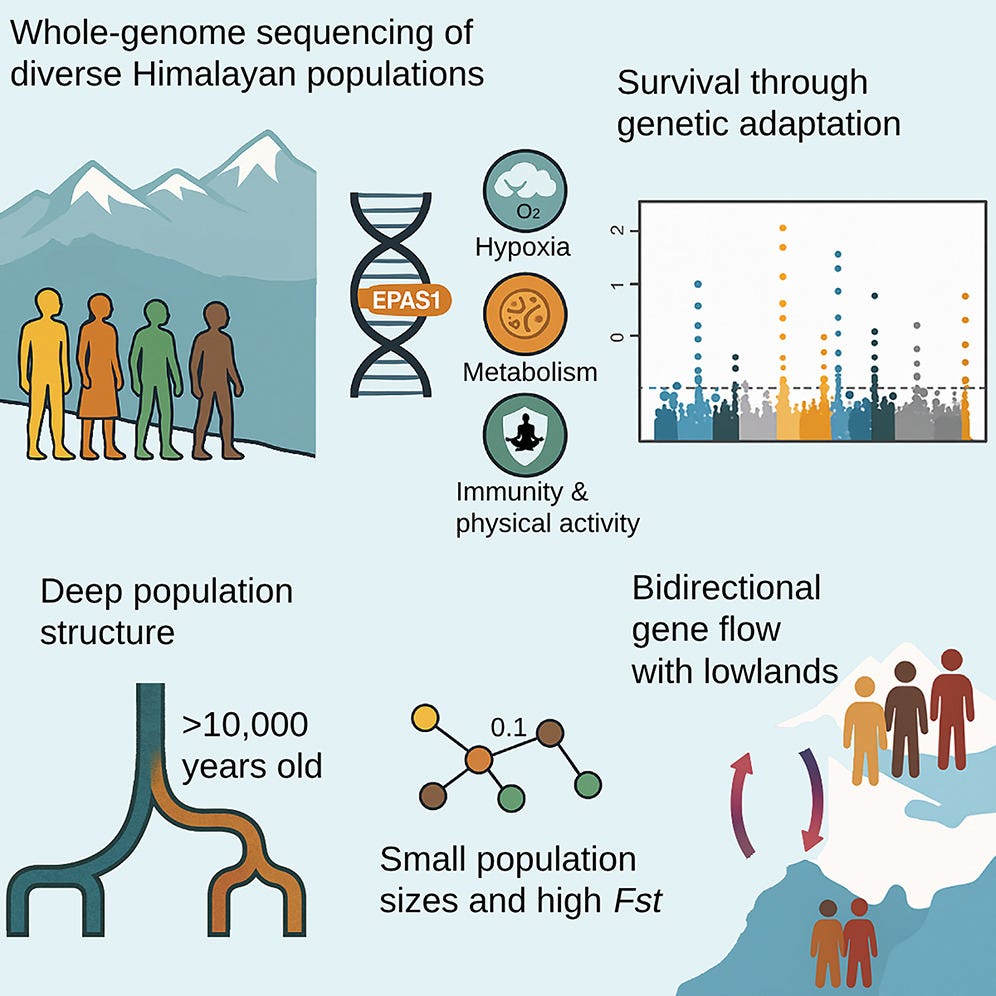
The Himalayas have long stood as a boundary between empires, languages, and ecologies. Yet even as a formidable barrier, these mountains have hosted human life for millennia. A new genomic study, published in Current Biology1 by Arciero et al. (2025), offers a sweeping view of the genetic legacy of Himalayan populations. It is not just a study of altitude survival, but one that reframes how scientists understand the early peopling, isolation, and biological resilience of communities in some of the harshest habitats on Earth.
By sequencing whole genomes from 87 individuals across 16 ethnic groups in Bhutan, Nepal, India, and the Tibetan plateau, researchers identified a complex population history that stretches back more than 10,000 years. These lineages predate permanent highland settlements by several millennia. In the words of the authors:
"The deep population structure observed across Himalayan groups indicates early divergence well before archaeological signs of sedentary life above 2,500 meters."
These results challenge assumptions that permanent high-altitude living began only with agriculture. Instead, genetic footprints suggest that small, isolated groups adapted to hypoxic conditions thousands of years earlier, likely through sustained presence at moderate elevations.
Echoes of Archaic Ancestors
One of the study’s key findings centers on the EPAS1 gene—often called the "super athlete gene"—which regulates the body’s response to low oxygen. This gene, inherited from Denisovans, an extinct archaic human species, has long been associated with Tibetan highlanders.
In this broader Himalayan dataset, however, EPAS1 was found at high frequencies across all high-altitude groups, including populations in Bhutan and eastern Nepal. The authors estimate that the introgressed version of the gene coalesced between 30,000 and 50,000 years ago:
"These Denisovan haplotypes were not only retained, but strongly selected in multiple populations, suggesting convergent adaptation to highland living across different parts of the Himalayas."
Other introgressed segments—some also found in Oceanian populations—were identified in regions of the genome related to immune regulation and blood pH control, hinting at shared ancient pressures tied to hypoxia and diet.
Genetic Isolation and the Weight of Mountains
Perhaps more surprising than shared adaptation was the degree of divergence between Himalayan groups. Using a battery of genomic tools, the team showed that many of these populations—like the Lhokpu in Bhutan and the Toto in India—have some of the highest levels of homozygosity in the world. Their genomes reflect long-term isolation and repeated bottlenecks:
"FST values between some Himalayan groups rival those seen between populations on different continents."
High genetic drift and limited gene flow shaped these differences, reinforced by geography and social boundaries. Populations often remained separated by only a few mountain valleys yet show substantial genomic distinctiveness.
A Corridor, Not a Wall
Still, the Himalayas were not impermeable. The study detected waves of gene flow both into and out of the highlands. Between 800 and 450 CE, for example, Tibetan expansion appears to have brought Himalayan ancestry westward into Central Asian groups like the Burusho. Meanwhile, gene flow from northern India moved up into Nepal, likely during the era of the Gupta Empire.
This exchange was not symmetrical. In groups such as the Chetri, researchers found evidence of sex-biased admixture, with a stronger signal of South Asian male ancestry. In contrast, the Tharu population seems to have received more balanced gene flow from both sexes.
The Totopara Enigma
Among the most curious findings is the presence of a Denisovan-derived region in the genomes of the Toto, a small and historically isolated group in northern India. This same region—located in the FAM178B gene—also shows up at high frequency in Oceanian populations and is linked to breath-hold diving among the Bajau of Indonesia.
Yet the Toto now live at low altitudes. How this adaptation made its way into their genome is unclear. One possibility is historical high-altitude residence followed by resettlement. Another is gene flow from East Asian groups where the variant remains common.
A Wider View on High-Altitude Genetics
Genome-wide scans for natural selection identified a suite of genes likely involved in adaptation to cold, oxygen scarcity, immune defense, and metabolism. These include variants in:
TBC1D1, involved in glucose and lipid metabolism
CLTCL1, regulating insulin response
PAX5 and VTCN1, immune-related pathways
SYNPO2L, associated with heart muscle function
Some of these genes are also under selection in cold-adapted animal species. Taken together, they paint a picture of a multigenic response to one of Earth’s most extreme climates.
The Cultural Significance of Genomic Histories
While the scientific focus is on molecular evolution, the study’s implications reach far into cultural anthropology. The preservation of distinct genetic signatures across small, often marginalized communities illustrates how tradition, language, and geography intersect with biology.
As noted by co-author Dr. Marc Haber:
"This work expands the lens through which we view Himalayan diversity—not just through archaeological fragments or linguistic trees, but through the very code carried by living people."
By anchoring genomic signals to archaeological timelines and historical migrations, this research provides a more integrated framework for understanding the long and intricate story of Himalayan settlement.
A Future for High-Altitude Genomics
The authors hope this research encourages further collaboration with underrepresented populations. Many of the groups in this study had not been previously sampled at a genomic level. Including them provides critical context and corrects biases inherent in global datasets.
Future work may explore how these adaptive variants affect modern health outcomes in highland populations, or how ancient adaptations interact with new challenges such as climate change.
In a region where landscapes seem frozen in time, these genomes show a history in motion: one of survival, movement, separation, and adaptation—written in the blood and bones of people who have lived among the clouds.
Additional Related Research
Huerta-Sánchez, E., Jin, X., Asan, Bianba, Z., Peter, B. M., Vinckenbosch, N., Yi, X., ... & Nielsen, R. (2014). Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature, 512(7513), 194–197. https://doi.org/10.1038/nature13408
Jeong, C., et al. (2016). Long-term genetic stability and a high-altitude East Asian origin for the peoples of the high valleys of the Himalayan arc. Proceedings of the National Academy of Sciences, 113(27), 7485–7490. https://doi.org/10.1073/pnas.1520844113
Wang, H., et al. (2023). Human genetic history on the Tibetan Plateau in the past 5100 years. Science Advances, 9(5), eadd5582. https://doi.org/10.1126/sciadv.add5582
Liu, C. C., et al. (2022). Ancient genomes from the Himalayas illuminate the genetic history of Tibetans and their Tibeto-Burman speaking neighbors. Nature Communications, 13(1), 1203. https://doi.org/10.1038/s41467-022-28827-2
Arciero, E., et al. (2018). Demographic history and genetic adaptation in the Himalayan region inferred from genome-wide SNP genotypes of 49 populations. Molecular Biology and Evolution, 35(8), 1916–1933. https://doi.org/10.1093/molbev/msy094
Arciero, E., Almarri, M. A., Mezzavilla, M., Xue, Y., Hallast, P., Hammoud, C., Chen, Y., Skov, L., Kraaijenbrink, T., Ayub, Q., Yang, H., van Driem, G., Jobling, M. A., de Knijff, P., Tyler-Smith, C., Asan, & Haber, M. (2025). Whole-genome sequences provide insights into the formation and adaptation of human populations in the Himalayas. Current Biology, 35, 1-14. https://doi.org/10.1016/j.cub.2025.06.048