A Deep Time Archive of Infection
Until recently, most of what was known about ancient disease came from indirect traces: pitting in skeletal bones, ancient texts, or lingering scars in the archaeological record. But with the latest genomic tools, bones are becoming archives in their own right—records of ancient immune battles written in microbial DNA.
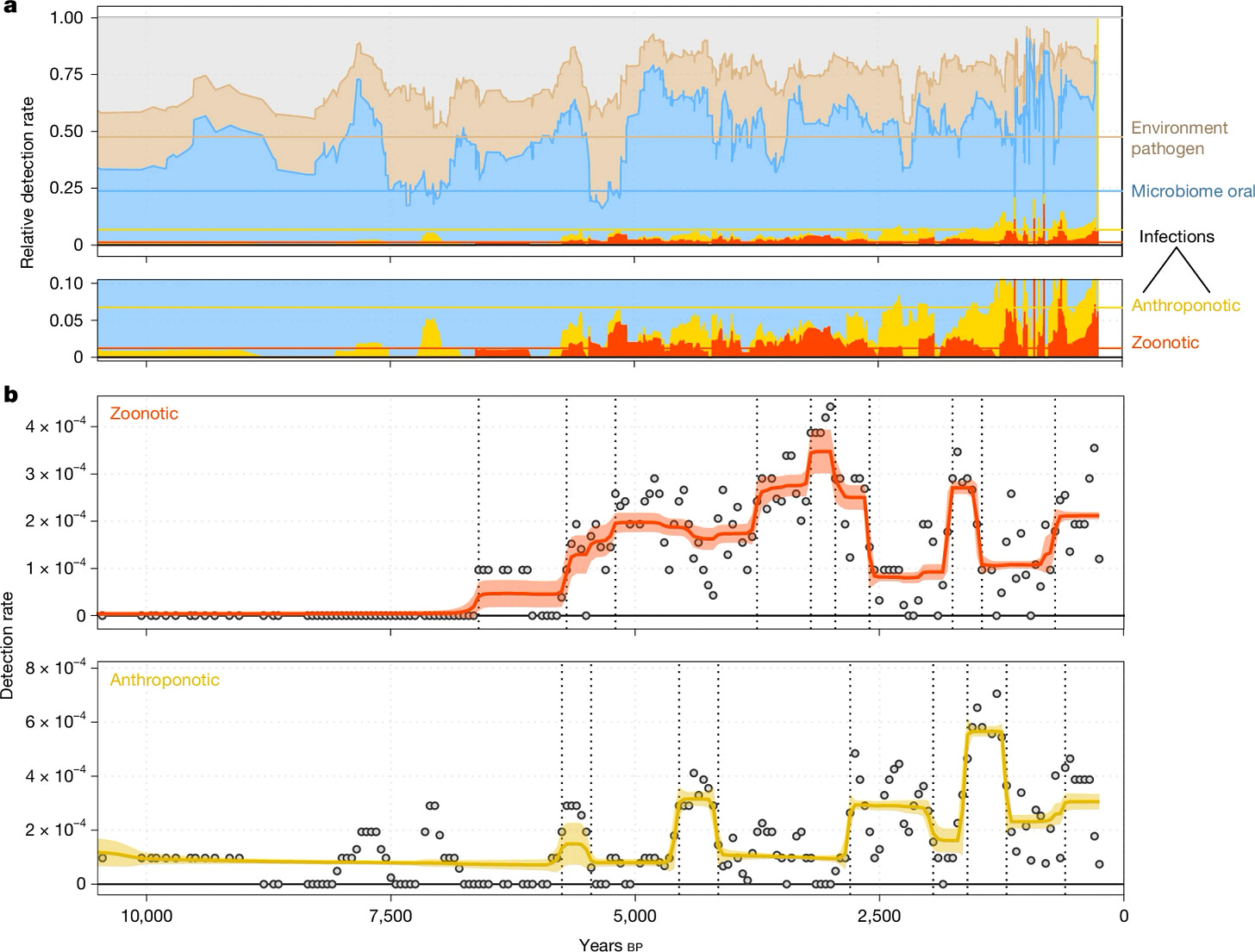
In a new study published in Nature1, a team led by Eske Willerslev of the University of Copenhagen and University of Cambridge reports the most extensive ancient pathogen survey ever conducted. Drawing from more than 1,300 prehistoric individuals across Eurasia, the researchers recovered DNA traces from 214 different pathogens—including the oldest known genetic fingerprint of the plague bacterium, Yersinia pestis, in a human who died 5,500 years ago.
"We've long suspected that the transition to farming and animal husbandry opened the door to a new era of disease. Now DNA shows us that it happened at least 6,500 years ago," said Willerslev.

The findings suggest a pivotal shift: as early farmers settled down, built animal pens, and traveled with their herds, they also created the perfect conditions for pathogens to leap from animals to humans.
How Domestication Helped Shape Human Disease
For most of human evolution, small groups of mobile foragers kept their distance from each other and from concentrated sources of contagion. But that changed with agriculture. Around 10,000 years ago, in the Fertile Crescent and surrounding regions, people began living in close quarters—not just with one another, but with domesticated pigs, sheep, cattle, and goats.

This new proximity set the stage for zoonoses: diseases that spread from animals to humans. Among the 214 pathogens identified in the study were multiple bacteria, viruses, and parasites known to infect both livestock and people. According to the study, the earliest widespread appearance of these zoonoses occurred around 6,500 years ago.
"Mutations that were successful in the past are likely to reappear. This knowledge is important for future vaccines," said Martin Sikora, co-author of the study. "It allows us to test whether current vaccines provide sufficient coverage—or whether new ones need to be developed due to mutations."
The genetic data aligns with the archaeological record showing growing complexity in settlement patterns, trade routes, and social stratification during the Neolithic and Bronze Age. As people began to move in larger numbers—especially during the steppe migrations from regions like the Pontic-Caspian steppe—so did their pathogens.
The World's Oldest Plague
Among the study’s most striking discoveries is the earliest genetic evidence of Yersinia pestis, the bacterium that causes plague. Previously, the oldest known sample of Y. pestis came from roughly 5,000 years ago in Central Asia. The new record, dated to 5,500 years ago, pushes the timeline further back and strengthens the link between agricultural transitions and the emergence of epidemic disease.
Plague would later devastate medieval Europe during the Black Death, but its roots go deeper—into the soil of early farming villages, where rodents and their fleas found new ecological niches. This reinforces the idea that some of humanity's most catastrophic diseases were not born during times of collapse, but times of growth.
Disease as a Driver of Prehistoric Change
The consequences of this microbial shift were not limited to health. The researchers suggest that ancient diseases may have contributed to population decline, mobility, and even evolutionary change. Just as modern pandemics have reshaped politics and economies, prehistoric pandemics may have influenced migration, warfare, and demographic replacement.
"These infections didn't just cause illness. They may have contributed to population collapse, migration, and genetic adaptation," said Willerslev.
The genomic data also support the idea that natural selection acted strongly on immune-related genes during the Neolithic and Bronze Ages. Genes associated with inflammation, antiviral defense, and immunity appear to have been under strong pressure, hinting at waves of infection reshaping the human gene pool.
What the Past Means for the Future
Beyond its historical implications, this study offers a roadmap for anticipating future health challenges. The team’s genetic reconstructions provide clues about how pathogens evolve, what kinds of mutations tend to recur, and which immune responses are most frequently targeted.
By studying how early farming societies responded to microbial threats, researchers can better understand the vulnerabilities of modern populations as they confront new zoonoses in an era of industrial-scale animal farming, global trade, and climate change.
Related Research
For readers interested in the genetic evolution of disease and its long-term impacts, several recent studies complement this work:
Rasmussen, S., et al. (2015). Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell, 163(3), 571–582. https://doi.org/10.1016/j.cell.2015.10.009
Spyrou, M. A., et al. (2018). Historical Yersinia pestis genomes reveal the European Black Death as the origin of ancient and modern plague pandemics. Cell Host & Microbe, 24(6), 814–821. https://doi.org/10.1016/j.chom.2018.10.004
Andrades Valtueña, A., et al. (2022). Stone Age Yersinia pestis genomes shed light on the spread and evolution of plague. Nature, 606(7912), 885–891. https://doi.org/10.1038/s41586-022-04721-5
Sikora, M., Canteri, E., Fernandez-Guerra, A., Oskolkov, N., Ågren, R., Hansson, L., Irving-Pease, E. K., Mühlemann, B., Holtsmark Nielsen, S., Scorrano, G., Allentoft, M. E., Valeur Seersholm, F., Schroeder, H., Gaunitz, C., Stenderup, J., Vinner, L., Jones, T. C., Nystedt, B., Sjögren, K.-G., … Willerslev, E. (2025). The spatiotemporal distribution of human pathogens in ancient Eurasia. Nature, 1–9. https://doi.org/10.1038/s41586-025-09192-8