In a quiet room humming with server stacks, a genomic dataset from nearly 300,000 Americans is doing something anthropologists have long tried to accomplish: capturing a living mosaic of human ancestry at a scale once unimaginable.
The participants belong to the All of Us Research Program, an ambitious effort by the National Institutes of Health to create a biomedical database that actually reflects the population of the United States—not just those whose ancestry traces back to Northern Europe.
The result1? A genomic map layered not only with centuries of migrations and admixture, but also with lessons for how ancestry—understood through the language of DNA—shapes and is shaped by human history, cultural geography, and inequality.
“What emerges is a complex web of similarity clusters,” notes the study’s senior author I. King Jordan. “They reflect not just where people come from, but how populations have blended, diverged, and lived together over time.”
Clusters Within Clusters
The researchers used principal component analysis (PCA) and a rapid ancestry estimation method called Rye to identify distinct patterns of population structure. This wasn’t simply a technical exercise. Their findings showed clusters of closely related individuals embedded in a much broader tapestry of admixture. Seven major clusters emerged from PCA, but further analysis using another method (UMAP) revealed up to 13 groupings—suggesting layers of population complexity.
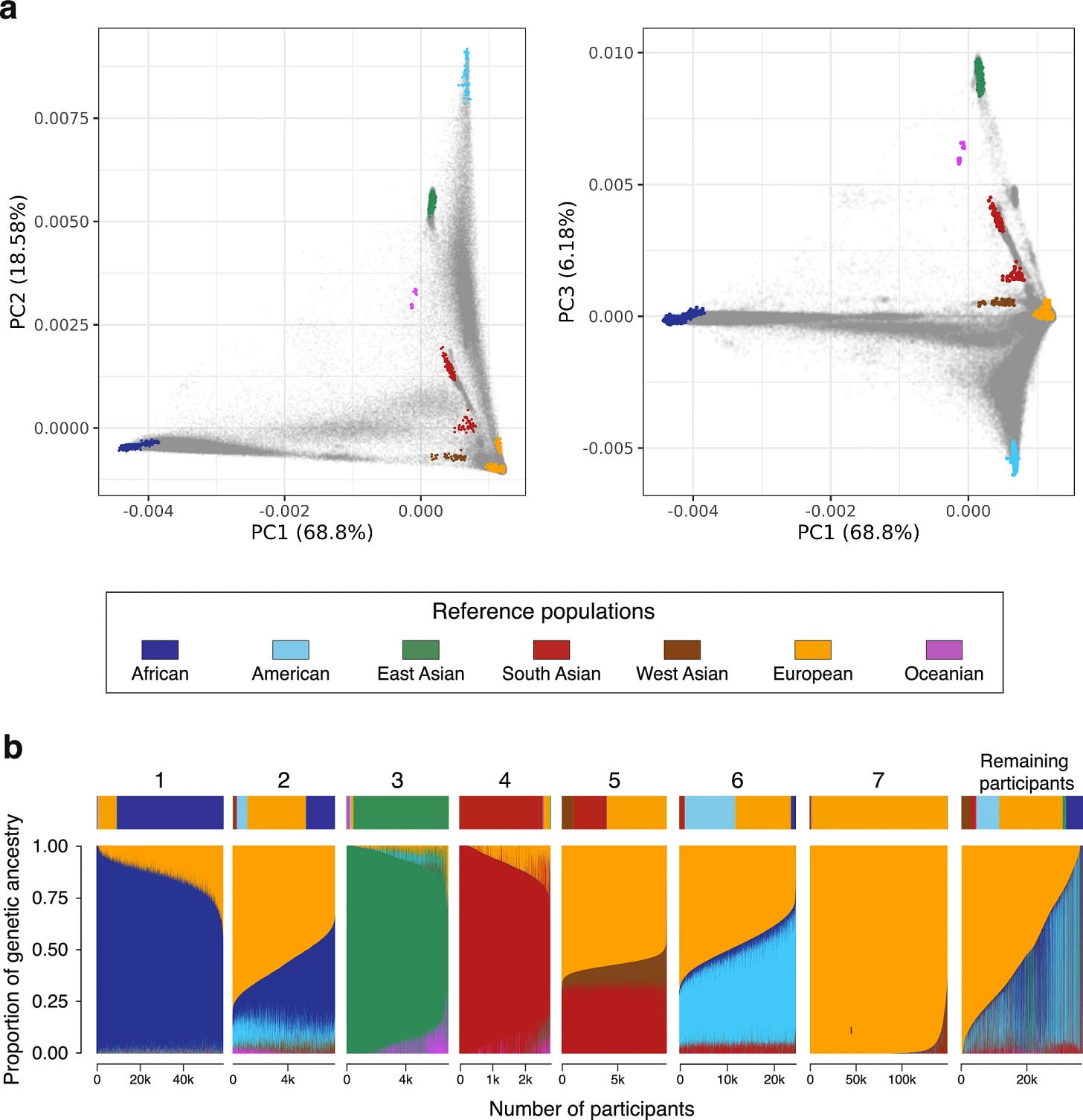
While the majority of participants had significant European ancestry (~66.4%), almost 20% showed African ancestry, and over 13% carried lineages from Indigenous American, South Asian, East Asian, or West Asian origins.
“We’re not just seeing neat little bins of ancestry,” said Jordan. “We’re seeing gradients, overlaps, mosaics—and that’s important because this is how real human populations exist.”
This granular structure offers more than genealogical trivia. It challenges how researchers traditionally categorize populations. Many of these ancestry components do not align cleanly with socially constructed racial or ethnic identities, exposing the limits of fixed labels in science, medicine, and society.
Admixture as a Time Machine
Genetic ancestry doesn’t just map where someone’s relatives lived—it can also hint at when they lived and migrated. The study revealed a striking generational pattern: younger Americans tend to be more genetically admixed than their elders.
In other words, the country’s population isn’t just becoming more diverse—it’s becoming more genomically entangled.
This has implications beyond genomics. It speaks to cultural fluidity, the erosion of categorical boundaries, and the demographic reality that human identity has always been more braided than boxed.
Geography of DNA
The research team also mapped these ancestry components across all 50 states and Puerto Rico. Predictably, African ancestry is most concentrated in the southeastern United States, while Indigenous American ancestry is more prominent in the Southwest and in California. European ancestry dominates throughout but shows the highest concentration in the northern Midwest and New England.
Importantly, the northeast corridor and Florida showed higher levels of admixture, highlighting urban regions as mixing zones for human histories.
“This isn’t just about ancestry,” explains co-author Robert Meller. “It’s about how geography, migration, and policy have shaped—and continue to shape—what it means to be American.”
Archaeogenetics for the Living
Although typically the domain of ancient DNA, this study demonstrates how archaeological thinking—about movement, kinship, and cultural entanglement—can be applied to contemporary populations.
It also underscores the limitations of using a Eurocentric reference framework. For about 3% of the participants, especially those with South or West Asian ancestry, ancestry estimates were sensitive to the choice of reference populations. This is a call to action: geneticists need to expand and diversify their reference datasets, much like how archaeologists must question whose stories get preserved in the record.
The team is aware of these pitfalls.
“Reference populations are idealized,” said Jordan. “They represent snapshots in time, often shaped by modern national boundaries or linguistic identities, which don’t always reflect deeper historical processes.”
A Different Kind of Deep Time
Anthropologists often work with bones, tools, and ancient genomes to tell the story of our species. But here, in the swabbed cheeks and sequenced blood of nearly 300,000 living people, another archive emerges—one that extends deep into the past but remains entirely alive.
The All of Us cohort is both a mirror and a time capsule. It shows how genetic variation echoes ancient human movements, colonial entanglements, the transatlantic slave trade, Indigenous resilience, and waves of global migration—all the while pointing to the future of a society where genetic boundaries are increasingly hard to draw.
A Note on the Future
This project is only the beginning. With every new genome added, the picture sharpens. But so does the need to think critically about how we use ancestry labels, who gets represented in the data, and what stories we choose to tell.
Precision medicine can’t be truly “precise” if it leaves out large swaths of humanity. This research is a reminder that the diversity of the human story must be matched by the diversity of our science.
Related Research
Abul-Husn, N. S., & Kenny, E. E. (2019). Personalized medicine and the power of electronic health records. Cell, 177(1), 58–69. https://doi.org/10.1016/j.cell.2019.02.039
Hellenthal, G., et al. (2014). A genetic atlas of human admixture history. Science, 343(6172), 747–751. https://doi.org/10.1126/science.1243518
Maples, B. K., et al. (2013). RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet., 93(2), 278–288. https://doi.org/10.1016/j.ajhg.2013.06.020
Bryc, K., et al. (2015). The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet., 96(1), 37–53. https://doi.org/10.1016/j.ajhg.2014.11.010
Sharma, S., Nagar, S. D., Pemu, P., Zuchner, S., SEEC Consortium, Mariño-Ramírez, L., Meller, R., & Jordan, I. K. (2025). Genetic ancestry and population structure in the All of Us Research Program cohort. Nature Communications, 16(1), 4123. https://doi.org/10.1038/s41467-025-59351-8