What makes the human brain so different from that of our closest relatives? The answer is not found in a single gene but in stretches of DNA that regulate when and how other genes turn on. A new study from the University of California San Diego identifies one such region, a molecular “switch” called HAR123, that may have tilted our evolutionary trajectory toward greater flexibility of thought.
The findings, published in Science Advances1, suggest that HAR123 influenced how our developing brains balanced the production of neurons and glial cells, two fundamental building blocks of neural circuits. That balance, in turn, may have opened the door to cognitive adaptability, the capacity to revise old information and learn new rules—a trait that sets humans apart from other primates.
“HAR123 is not a gene in itself, but a powerful enhancer that shapes the activity of other genes during brain development,” said senior author Miles Wilkinson, UC San Diego School of Medicine.
A Region That Evolved in the Fast Lane
HAR123 belongs to a class of DNA sequences known as human-accelerated regions (HARs). These are parts of the genome that remained largely unchanged across millions of years of mammalian evolution, only to undergo a burst of rapid mutations after the lineage leading to humans split from that of chimpanzees roughly 5 to 6 million years ago.
That burst of change suggests selective pressures were at play. Instead of drifting randomly, HARs likely carried alterations that offered advantages in cognition, development, or adaptation. In the case of HAR123, the research team found that its human-specific sequence drove stronger activity than the chimpanzee version when tested in neural progenitor cells.
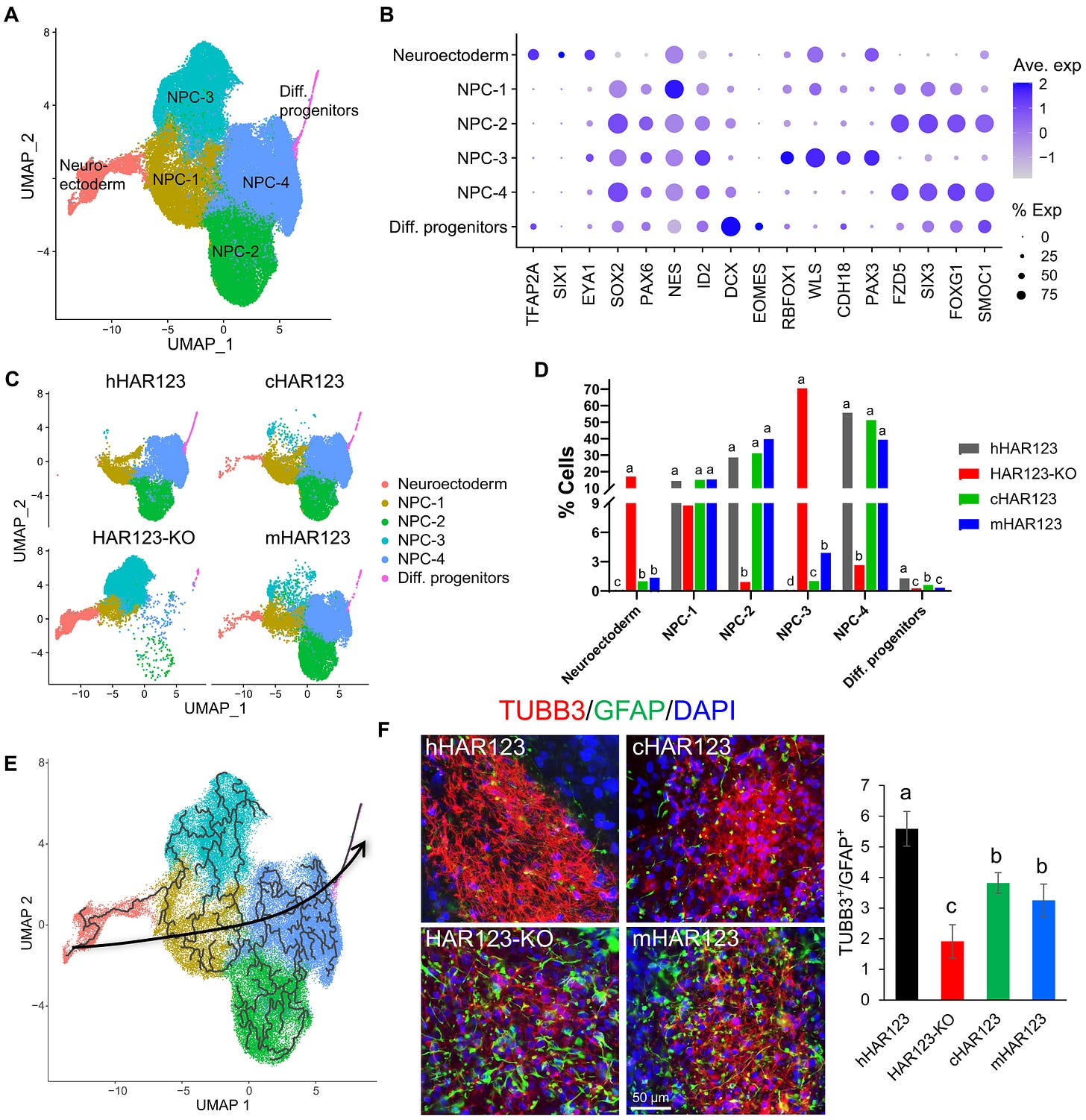
These progenitor cells are the stem-like precursors that generate both neurons, responsible for processing information, and glia, which support and regulate neural networks. By tweaking how many of each type are produced, HAR123 may have fine-tuned the architecture of the human brain.
Cognitive Flexibility and the Human Condition
The study suggests HAR123 promoted an especially human trait: cognitive flexibility. This is the ability to discard outdated knowledge, adopt new strategies, and switch between modes of thinking. Archaeological records, from the appearance of new stone tool traditions to shifts in symbolic expression, reflect the importance of this trait throughout human prehistory.
“Our data suggest HAR123 may have contributed to the neural substrate that makes cognitive flexibility possible,” said co-author Kun Tan.
When the human and chimpanzee versions of HAR123 were compared in lab-grown stem cells, the enhancer shaped gene activity differently in each case. The human version promoted more dynamic regulation of neural progenitors, potentially granting our brains a broader repertoire of cell types and connections.
From Evolution to Disorder
The same molecular pathways that fueled the rise of human cognition may also underlie certain vulnerabilities. The authors note that HAR123, and HARs more broadly, are linked to neurodevelopmental conditions such as autism. While HAR123’s human-specific changes may have facilitated adaptability, they may also have introduced susceptibilities that our species continues to live with today.
Why This Matters for Human Origins
For anthropologists and archaeologists, HAR123 provides a molecular foothold for one of the most elusive questions in human evolution: how small tweaks in DNA rewired the brain. Unlike fossils or artifacts, enhancers such as HAR123 do not leave visible traces in the ground. Yet they may explain why our ancestors were capable of innovation, from Acheulean handaxes to Upper Paleolithic cave art.
In this sense, HAR123 bridges the molecular and the cultural, linking mutations in a regulatory sequence to the behaviors that define our lineage.
Related Research
Pollard, K. S., et al. (2006). “An RNA gene expressed during cortical development evolved rapidly in humans.” Nature, 443(7108), 167–172. https://doi.org/10.1038/nature05113
Hubisz, M. J., & Pollard, K. S. (2014). “Exploring the genesis and functions of Human Accelerated Regions sheds light on their role in human evolution.” Current Opinion in Genetics & Development, 29, 15–21. https://doi.org/10.1016/j.gde.2014.07.005
Doan, R. N., et al. (2016). “Mutations in human accelerated regions disrupt cognition and social behavior.” Cell, 167(2), 341–354.e12. https://doi.org/10.1016/j.cell.2016.08.071
Capra, J. A., et al. (2013). “Many human accelerated regions are developmental enhancers.” Philosophical Transactions of the Royal Society B, 368(1632), 20130025. https://doi.org/10.1098/rstb.2013.0025
Tan, K., Higgins, K., Liu, Q., & Wilkinson, M. F. (2025). An ancient enhancer rapidly evolving in the human lineage promotes neural development and cognitive flexibility. Science Advances, 11(33), eadt0534. https://doi.org/10.1126/sciadv.adt0534